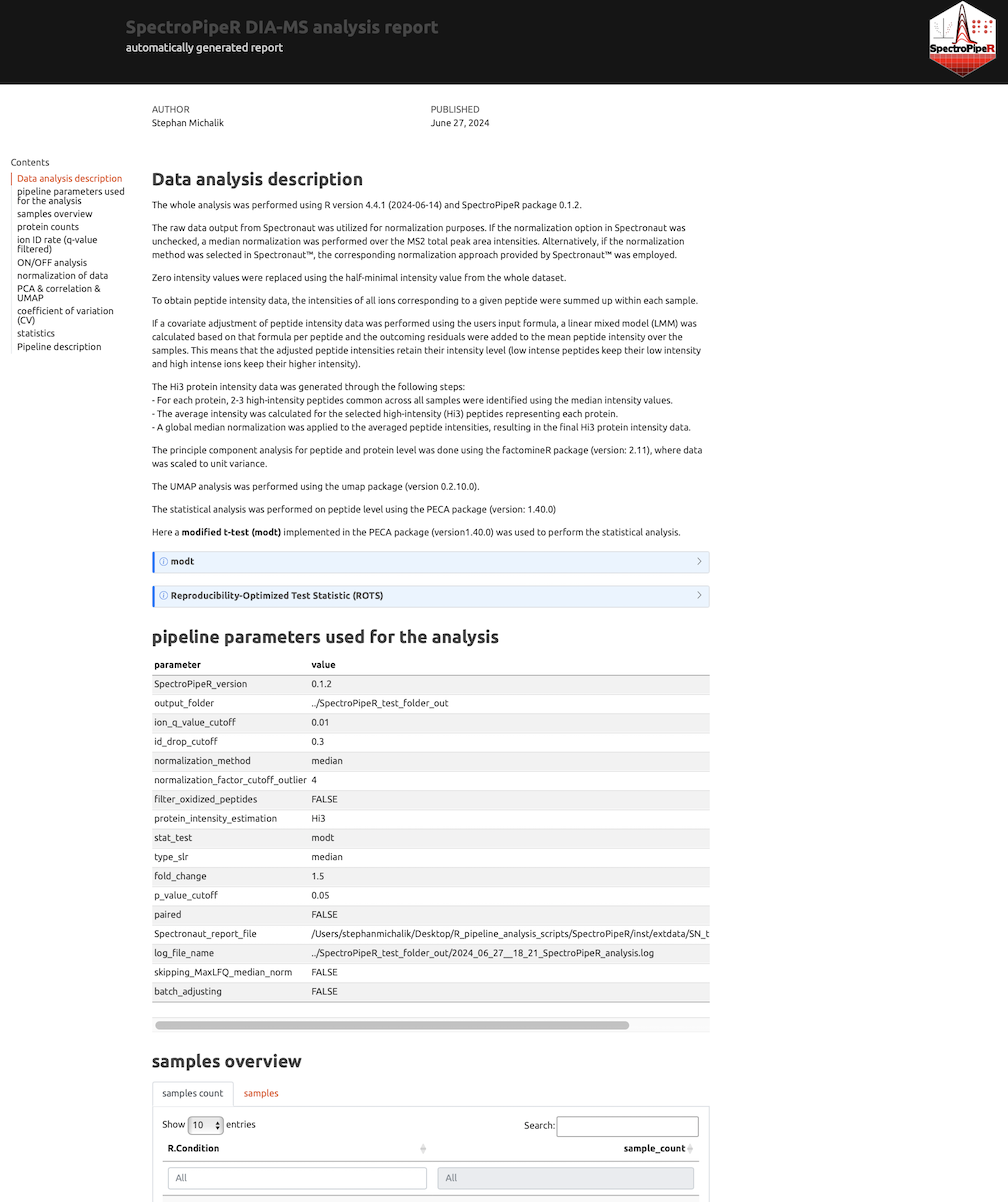
SpectroPipeR - step 5 - reporting
a05_SpectroPipeR_reporting.RmdSpectroPipeR reporting
The reporting module takes the inputs from:
- step 1 - read Spectronaut data
- step 2 - normalization and quantification
- step 5 - statistics (optional)
to render an interactive standalone HTML report. The rendering is performed with Quarto CLI. So Quarto CLI is required. If you did not already install the Quarto CLI you should install the Quarto CLI using Quarto get started installation.
SpectroPipeR functions executed before
# parameter list
params <- list(output_folder = "../SpectroPipeR_test_folder")
# example input file, bundled with SpectroPipeR package
example_file_path <- system.file("extdata", "SN_test_HYE_mix_file.tsv", package="SpectroPipeR")
# step 1: load Spectronaut data module
SpectroPipeR_data <- read_spectronaut_module(file = example_file_path,
parameter = params,
print.plot = FALSE)
# step 2: normalize & quantification module
SpectroPipeR_data_quant <- norm_quant_module(SpectroPipeR_data = SpectroPipeR_data,print.plot = FALSE)
# step 3: MVA module
MVA_module(SpectroPipeR_data_quant = SpectroPipeR_data_quant)
# step 4: statistics module
SpectroPipeR_data_stats <- statistics_module(SpectroPipeR_data_quant = SpectroPipeR_data_quant,
condition_comparisons = cbind(c("B_manual","A_manual")))Report generation
# step 5: report module
SpectroPipeR_report_module(SpectroPipeR_data = SpectroPipeR_data,
SpectroPipeR_data_quant = SpectroPipeR_data_quant,
SpectroPipeR_data_stats = SpectroPipeR_data_stats)
# #*****************************************
# # REPORT MODULE
# #*****************************************
#
# generating methods part ...
# render HTML report ... this might take a while
#
# processing file: DIA_MS_analysis_report_Master.qmd
# |............... | 30% # A tibble: 1 × 1
# value
# <chr>
# 1 0.01
#
# output file: DIA_MS_analysis_report_Master.knit.md
#
# pandoc --output SpectroPipeR_report.html
# to: html
# standalone: true
# self-contained: true
# section-divs: true
# html-math-method: katex
# wrap: none
# default-image-extension: png
# css:
# - styles.css
# toc: true
# toc-depth: 3
#
# metadata
# document-css: false
# link-citations: true
# date-format: long
# lang: en
# title: SpectroPipeR DIA-MS analysis report
# author: Stephan Michalik
# date: '`r format(Sys.Date(), "%B %d, %Y")`'
# title-block-banner: '#151515'
# subtitle: automatically generated report
# page-layout: full
# toc-title: Contents
# theme: united
# highlight: tango
# df_print: paged
# toc-location: left
# anchor-sections: true
# smooth-scroll: true
#
# Output created: SpectroPipeR_report.html
#
# render HTML report ... DONE!